1. Group
of Fox used Tetrazine-trans-cyclooctene
reaction for creating diffusion controlled 3D patterns (put it simply: particles) that could be filled with cells. Not a surprise
cells stayed viable. The particles after all were made of hyaluronic acid
derivatized with bioorthogonal chemistry that potentially can’t cause a harm. This
work can find it’s future application in cell and tissue printing.
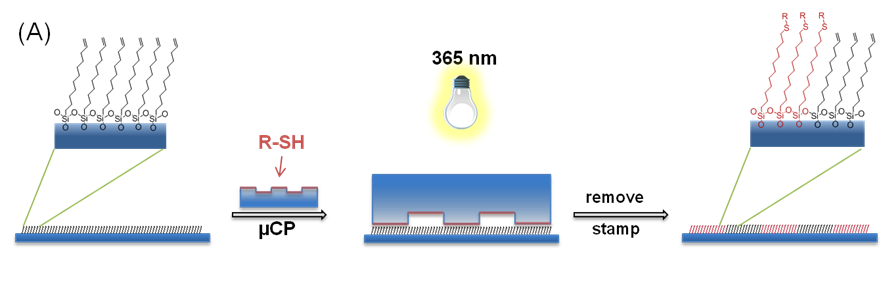
2. Conceptually
similar to the previous work, researchers in the group of Bart
Jan Ravoo printed set of chemical functionalities using inverse-electron demand
Diels-Alder chemistry (iEDDA) on the glass surfaces. In order to create those
they employed a microcontact chemistry which enables to create large area
patterns with high resolution. Essentially, you apply an ink that contain
chemicals that you want to print on the surface on the PDMS stamps of defined
geometry.
Next, you apply the stamp on the functionalised glass. The chemical coupling will
occur only at the sites where groups are close to each other. Very cool and straightforward.
I believe that this sort of chemistry will find its further development in the field
of biosensors and microfluidics as long as they fix a few limitations of this technique.
This are the low reproducibility of the PDMS stamp-patterning (due to its elasticity)
and a necessity to use apolar inks which is again due to the high hydrophobicity
of the PDMS.
3. A
biomolecule patterning is also a passion of Dr.
Hongyan Sun whi published her work in Chemical Communication son the
immobilization of peptides and proteins on surfaces with use of the iEDDA. A protein microarray technology may benefit from from this work.